My projects#
A showcase of my contributions to computational biology and genomics through various projects.
Deep Learning in Genomics#
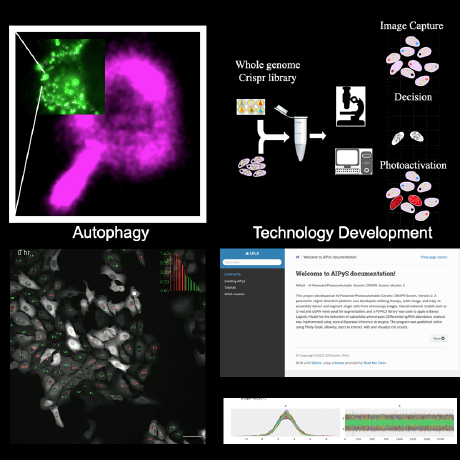
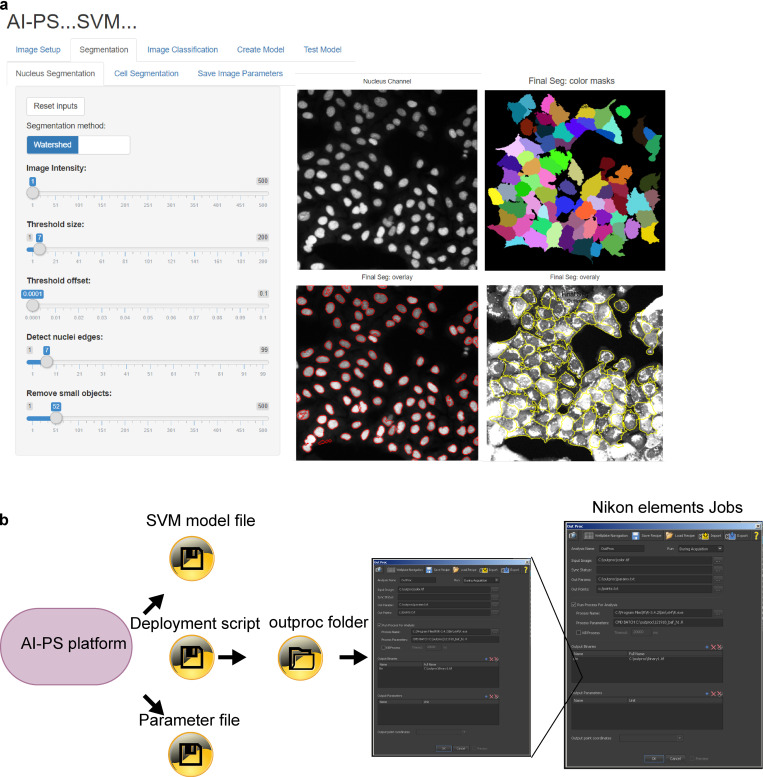
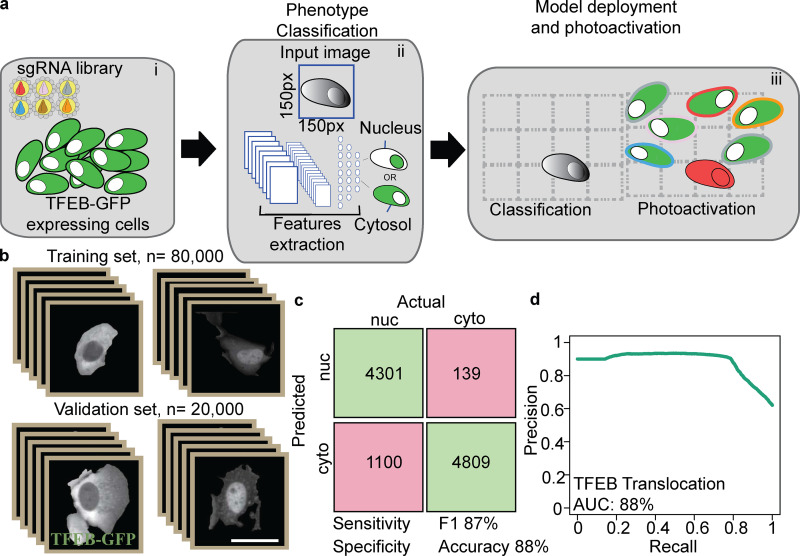
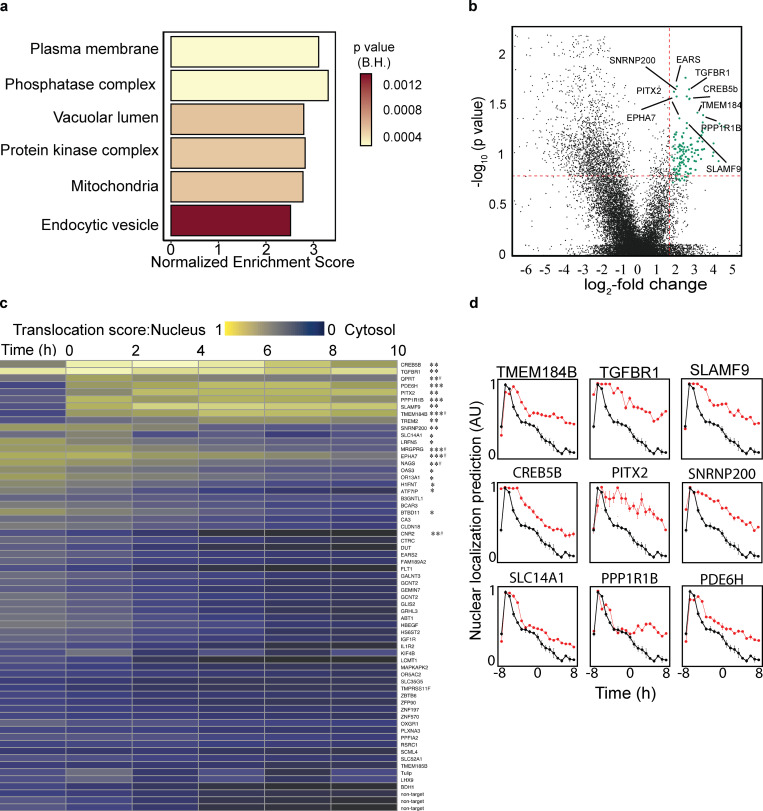
The AI Powered Photoswitchable Genetic CRISPR Screen, Version 2 (AIPyS) project represents a significant leap in applying parametric object detection in genomic research. By employing advanced deep learning models like U-net and cGAN within this initiative, I’ve facilitated object detection and the segmenting of genomic elements more precisely and efficiently. This project has been pivotal in accelerating functional genomic discoveries.
In the AI-Powered Photoswitchable Genetic CRISPR Screen, Version 1 (AIPSr), I developed a neural network-based cell classifier using Keras/TensorFlow. This project integrated deep learning techniques to revolutionize cell classification methods, enhancing the accuracy of genomic screenings significantly. Our deployment program integrated this platform with screening microscopes, pushing the frontiers of automated genomic analyses.
Genomic Data Analysis Tools#
I have developed an Automated Pipeline for Genomic Data Analysis, which streamlines the processing of transcriptomic data, particularly focusing on Parkinson’s disease research. This tool employs EdgeR and GSEA, integrated with Cystoscope for visual analytics, providing a comprehensive platform for functional enrichment analysis. This pipeline showcases my commitment to advancing biomedical research through the creation of highly efficient, automated analysis tools.
The project on Characterizing Protein-Protein Interactions involves in-depth proteomic analyses to understand the dynamics of protein interaction networks in dividing cells. Utilizing affinity purification coupled with SILAC, this research offers insights into the molecular mechanisms regulating cell division and highlights potential therapeutic targets for cellular dysfunction diseases. The application of this knowledge spans numerous fields, including cancer research and neurodegenerative disease studies.
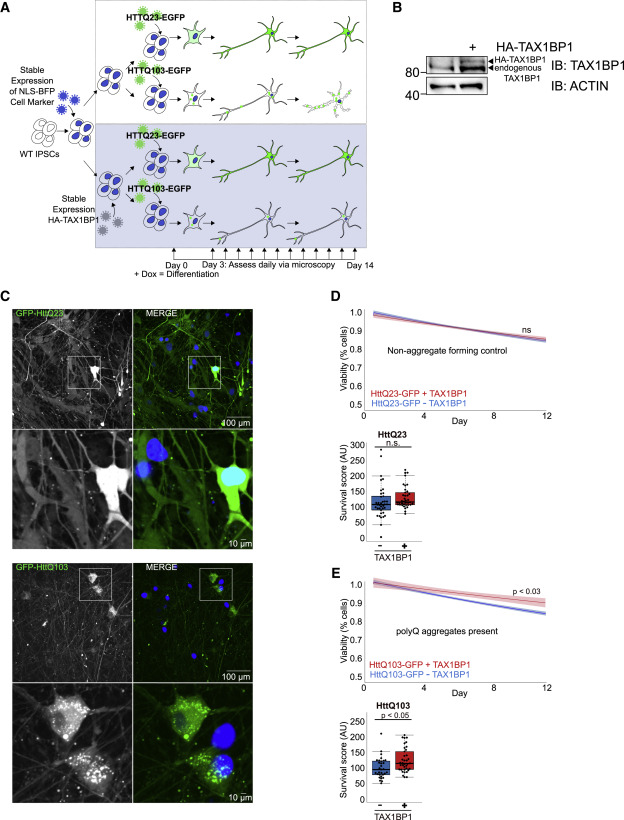
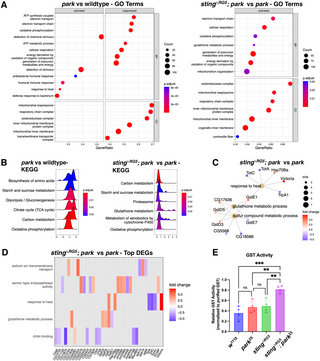
Here Project showcases my contributions to understanding Parkinson’s disease by creating and utilizing a custom pipeline for analyzing transcriptomic data from Parkin and STING gene mutants in animal models. This pipeline integrates bioinformatics methodologies to reveal insights into the genetic mechanisms of disease progression. Additionally, I’ve implemented a Gene Enrichment Analysis Pipeline to systematically identify significant pathways and gene functions that may play a role in Parkinson’s pathology. These tools have been instrumental in uncovering potential therapeutic targets. project showcases my contributions to understanding Parkinson’s disease by creating and utilizing a custom pipeline for analyzing transcriptomic data from Parkin and STING gene mutants in animal models. This pipeline integrates bioinformatics methodologies to reveal insights into the genetic mechanisms of disease progression. Additionally, I’ve implemented a Gene Enrichment Analysis Pipeline to systematically identify significant pathways and gene functions that may play a role in Parkinson’s pathology. These tools have been instrumental in uncovering potential therapeutic targets.
Expanding Localisation Microscopy Analysis#
Version 2 project is a monumental stride towards enhancing the analysis of localisation microscopy images. By incorporating sophisticated machine learning algorithms with high computing power, I’ve enabled faster and more accurate identification and subsequent analysis of microbial interactions. This toolkit has been instrumental in expediting breakthroughs in microorganism research and understanding of localisation microscopy.