parametric-segmentation
The AIPS packdge provides two alternative methods for segmenting cells: parametric or deep-learning segmentation. For parametric segmentation, we enhanced and translated our R-based code.
from AIPyS import AIPS_module as ai
from AIPyS import AIPS_functions as af
from AIPyS import AIPS_file_display as afd
Set AIPyS object for preforming segmentation of the nucleus (seed),
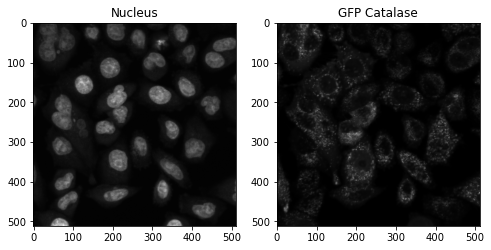
AIPS_object = ai.Segmentation(Image_name= 'catGFP.tif', path = 'data', ch_ = 1, rmv_object_nuc = 0.12,block_size = 59, offset=-4, clean = 3)
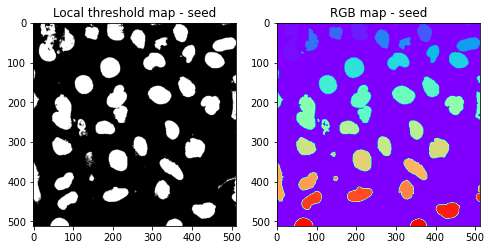
seed = AIPS_object.seedSegmentation()
nmask2 = seed['nmask2'] #Local threshold map - seed
sort_mask = seed['sort_mask'] #RGB map - seed
Then object is used for segmenting based on the nucleus as a seed.
where the seed segmentation parametrs are pluged in.
Calculate a threshold mask image by using a weighted mean (block_size) of the local neighborhood of each pixel, minus a offset.
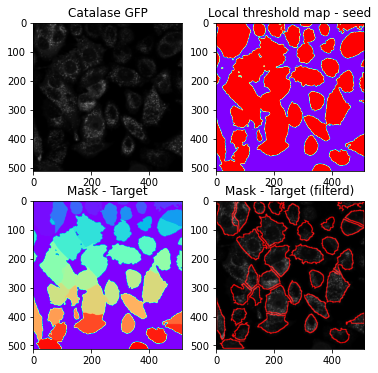
Target channel (Catalase-GFP) was used to identify cell borders and edges for segmentation. High-pass filtering, local thresholding, and global thresholding were then used to create global and local masks.
target = AIPS_object.cytosolSegmentation(ch2_=0, block_size_cyto=3, offset_cyto=-5, global_ther= 0.51, rmv_object_cyto=0.99, rmv_object_cyto_small=0.25)
combine = target['combine']
cseg_mask = target['cseg_mask']
mask_unfiltered = target['mask_unfiltered']
maskContour = afd.Compsite_display(input_image = image_pex[0,:,:], mask_roi = cseg_mask, channel= 0).draw_ROI_contour()